[Date Prev][Date Next][Thread Prev][Thread Next][Date Index][Thread Index]
[ccp4bb]: New ligand using Refmac5.0
Dear CCP4 users,
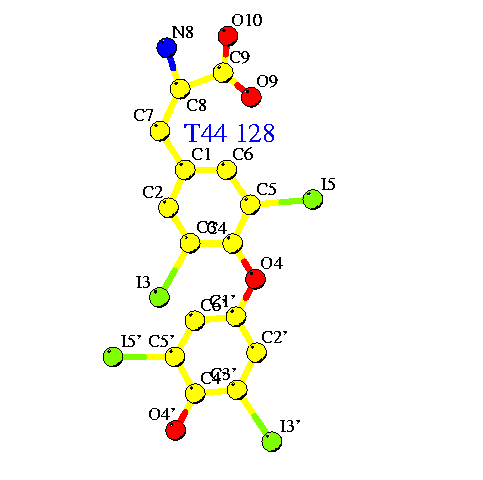
I am trying to refine a structure with 3,5,3',5'-tetraiodo-L-thyronine
(thyroxine, see attachment) using Refmac5.0.
I have downloaded the coordinates, built them into my structure, and let
refmac create a library describing the molecule, which looks like this:
global_
_lib_name mon_lib
_lib_version 3.1
_lib_update 16/07/00
# ------------------------------------------------
#
# --- LIST OF MONOMERS ---
#
data_comp_list
loop_
_chem_comp.id
_chem_comp.three_letter_code
_chem_comp.name
_chem_comp.group
_chem_comp.number_atoms_all
_chem_comp.number_atoms_nh
_chem_comp.desc_level
T44 T44 '3,5,3",5"-TETRAIODO-L-THYRONINE ' non-polymer
35 24 .
#
# --- DESCRIPTION OF MONOMERS ---
#
data_comp_T44
#
loop_
_chem_comp_atom.comp_id
_chem_comp_atom.atom_id
_chem_comp_atom.type_symbol
_chem_comp_atom.type_energy
_chem_comp_atom.partial_charge
T44 'I5'' I I 0.000
T44 'C5'' C CR6 0.000
T44 'C6'' C CR16 0.000
T44 'H6'' H HCR6 0.000
T44 'C4'' C CR6 0.000
T44 'O4'' O O 0.000
T44 'HO4'' H HO 0.000
T44 'C3'' C CR6 0.000
T44 'I3'' I I 0.000
T44 'C2'' C CR16 0.000
T44 'H2'' H HCR6 0.000
T44 'C1'' C CR6 0.000
T44 O4 O O 0.000
T44 C4 C CR6 0.000
T44 C5 C CR6 0.000
T44 I5 I I 0.000
T44 C6 C CR16 0.000
T44 H6 H HCR6 0.000
T44 C3 C CR6 0.000
T44 I3 I I 0.000
T44 C2 C CR16 0.000
T44 H2 H HCR6 0.000
T44 C1 C CR6 0.000
T44 C7 C CH2 0.000
T44 H71 H HCH2 0.000
T44 H72 H HCH2 0.000
T44 C8 C CH1 0.000
T44 H8 H HCH1 0.000
T44 N8 N N 0.000
T44 HN82 H HN 0.000
T44 HN81 H HN 0.000
T44 C9 C C 0.000
T44 O9 O O 0.000
T44 O10 O O 0.000
T44 HO1 H HO 0.000
loop_
_chem_comp_tree.comp_id
_chem_comp_tree.atom_id
_chem_comp_tree.atom_back
_chem_comp_tree.atom_forward
_chem_comp_tree.connect_type
T44 'I5'' n/a 'C5'' START
T44 'C5'' 'I5'' 'C4'' .
T44 'C6'' 'C5'' 'H6'' .
T44 'H6'' 'C6'' . .
T44 'C4'' 'C5'' 'C3'' .
T44 'O4'' 'C4'' 'HO4'' .
T44 'HO4'' 'O4'' . .
T44 'C3'' 'C4'' 'C2'' .
T44 'I3'' 'C3'' . .
T44 'C2'' 'C3'' 'C1'' .
T44 'H2'' 'C2'' . .
T44 'C1'' 'C2'' O4 .
T44 O4 'C1'' C4 .
T44 C4 O4 C3 .
T44 C5 C4 C6 .
T44 I5 C5 . .
T44 C6 C5 H6 .
T44 H6 C6 . .
T44 C3 C4 C2 .
T44 I3 C3 . .
T44 C2 C3 C1 .
T44 H2 C2 . .
T44 C1 C2 C7 .
T44 C7 C1 C8 .
T44 H71 C7 . .
T44 H72 C7 . .
T44 C8 C7 C9 .
T44 H8 C8 . .
T44 N8 C8 HN81 .
T44 HN82 N8 . .
T44 HN81 N8 . .
T44 C9 C8 O10 .
T44 O9 C9 . .
T44 O10 C9 HO1 .
T44 HO1 O10 . END
T44 C1 C6 . ADD
T44 'C1'' 'C6'' . ADD
loop_
_chem_comp_bond.comp_id
_chem_comp_bond.atom_id_1
_chem_comp_bond.atom_id_2
_chem_comp_bond.type
_chem_comp_bond.value_dist
_chem_comp_bond.value_dist_esd
T44 C1 C2 double 1.390 0.020
T44 C1 C6 single 1.390 0.020
T44 C7 C1 single 1.511 0.020
T44 C2 C3 single 1.390 0.020
T44 H2 C2 single 1.083 0.020
T44 C3 C4 double 1.487 0.020
T44 I3 C3 single 2.090 0.020
T44 C5 C4 single 1.487 0.020
T44 C4 O4 single 1.330 0.020
T44 C6 C5 double 1.390 0.020
T44 I5 C5 single 2.090 0.020
T44 H6 C6 single 1.083 0.020
T44 C8 C7 single 1.524 0.020
T44 H71 C7 single 1.092 0.020
T44 H72 C7 single 1.092 0.020
T44 C9 C8 single 1.500 0.020
T44 N8 C8 single 1.455 0.020
T44 H8 C8 single 1.099 0.020
T44 O9 C9 double 1.220 0.020
T44 O10 C9 single 1.330 0.020
T44 'C1'' 'C2'' double 1.390 0.020
T44 'C1'' 'C6'' single 1.390 0.020
T44 O4 'C1'' single 1.330 0.020
T44 'C2'' 'C3'' single 1.390 0.020
T44 'H2'' 'C2'' single 1.083 0.020
T44 'C3'' 'C4'' double 1.487 0.020
T44 'I3'' 'C3'' single 2.090 0.020
T44 'C4'' 'C5'' single 1.487 0.020
T44 'O4'' 'C4'' single 1.330 0.020
T44 'C6'' 'C5'' double 1.390 0.020
T44 'C5'' 'I5'' single 2.090 0.020
T44 'H6'' 'C6'' single 1.083 0.020
T44 HN81 N8 single 1.050 0.020
T44 HN82 N8 single 1.050 0.020
T44 'HO4'' 'O4'' single 0.890 0.020
T44 HO1 O10 single 0.890 0.020
loop_
_chem_comp_angle.comp_id
_chem_comp_angle.atom_id_1
_chem_comp_angle.atom_id_2
_chem_comp_angle.atom_id_3
_chem_comp_angle.value_angle
_chem_comp_angle.value_angle_esd
T44 'I5'' 'C5'' 'C6'' 120.000 3.000
T44 'I5'' 'C5'' 'C4'' 120.000 3.000
T44 'C6'' 'C5'' 'C4'' 120.000 3.000
T44 'C5'' 'C6'' 'H6'' 120.000 3.000
T44 'C5'' 'C6'' 'C1'' 120.000 3.000
T44 'H6'' 'C6'' 'C1'' 120.000 3.000
T44 'C5'' 'C4'' 'O4'' 120.000 3.000
T44 'C5'' 'C4'' 'C3'' 120.000 3.000
T44 'O4'' 'C4'' 'C3'' 120.000 3.000
T44 'C4'' 'O4'' 'HO4'' 120.000 3.000
T44 'C4'' 'C3'' 'I3'' 120.000 3.000
T44 'C4'' 'C3'' 'C2'' 120.000 3.000
T44 'I3'' 'C3'' 'C2'' 120.000 3.000
T44 'C3'' 'C2'' 'H2'' 120.000 3.000
T44 'C3'' 'C2'' 'C1'' 120.000 3.000
T44 'H2'' 'C2'' 'C1'' 120.000 3.000
T44 'C2'' 'C1'' O4 120.000 3.000
T44 'C2'' 'C1'' 'C6'' 120.000 3.000
T44 O4 'C1'' 'C6'' 120.000 3.000
T44 'C1'' O4 C4 120.000 3.000
T44 O4 C4 C5 120.000 3.000
T44 O4 C4 C3 120.000 3.000
T44 C5 C4 C3 120.000 3.000
T44 C4 C5 I5 120.000 3.000
T44 C4 C5 C6 120.000 3.000
T44 I5 C5 C6 120.000 3.000
T44 C5 C6 H6 120.000 3.000
T44 C5 C6 C1 120.000 3.000
T44 H6 C6 C1 120.000 3.000
T44 C4 C3 I3 120.000 3.000
T44 C4 C3 C2 120.000 3.000
T44 I3 C3 C2 120.000 3.000
T44 C3 C2 H2 120.000 3.000
T44 C3 C2 C1 120.000 3.000
T44 H2 C2 C1 120.000 3.000
T44 C2 C1 C7 120.000 3.000
T44 C2 C1 C6 120.000 3.000
T44 C7 C1 C6 120.000 3.000
T44 C1 C7 H71 109.470 3.000
T44 C1 C7 H72 109.470 3.000
T44 C1 C7 C8 109.470 3.000
T44 H71 C7 H72 107.900 3.000
T44 H71 C7 C8 109.470 3.000
T44 H72 C7 C8 109.470 3.000
T44 C7 C8 H8 108.340 3.000
T44 C7 C8 N8 105.000 3.000
T44 C7 C8 C9 109.470 3.000
T44 H8 C8 N8 109.470 3.000
T44 H8 C8 C9 108.810 3.000
T44 N8 C8 C9 111.600 3.000
T44 C8 N8 HN82 120.000 3.000
T44 C8 N8 HN81 120.000 3.000
T44 HN82 N8 HN81 120.000 3.000
T44 C8 C9 O9 120.500 3.000
T44 C8 C9 O10 120.500 3.000
T44 O9 C9 O10 120.000 3.000
T44 C9 O10 HO1 120.000 3.000
loop_
_chem_comp_tor.comp_id
_chem_comp_tor.id
_chem_comp_tor.atom_id_1
_chem_comp_tor.atom_id_2
_chem_comp_tor.atom_id_3
_chem_comp_tor.atom_id_4
_chem_comp_tor.value_angle
_chem_comp_tor.value_angle_esd
_chem_comp_tor.period
T44 CONST_1 'I5'' 'C5'' 'C6'' 'C1'' 180.000 0.000 0
T44 CONST_2 'I5'' 'C5'' 'C4'' 'C3'' 180.000 0.000 0
T44 var_1 'C5'' 'C4'' 'O4'' 'HO4'' 180.000 20.000 1
T44 CONST_3 'C5'' 'C4'' 'C3'' 'C2'' 0.000 0.000 0
T44 CONST_4 'C4'' 'C3'' 'C2'' 'C1'' 0.000 0.000 0
T44 CONST_5 'C3'' 'C2'' 'C1'' O4 180.000 0.000 0
T44 CONST_6 'C3'' 'C2'' 'C1'' 'C6'' 0.000 0.000 0
T44 var_2 'C2'' 'C1'' O4 C4 -9.718 20.000 1
T44 var_3 'C1'' O4 C4 C3 90.772 20.000 1
T44 CONST_7 O4 C4 C5 C6 180.000 0.000 0
T44 CONST_8 C4 C5 C6 C1 0.000 0.000 0
T44 CONST_9 O4 C4 C3 C2 180.000 0.000 0
T44 CONST_10 C4 C3 C2 C1 0.000 0.000 0
T44 CONST_11 C3 C2 C1 C7 180.000 0.000 0
T44 CONST_12 C3 C2 C1 C6 0.000 0.000 0
T44 var_4 C2 C1 C7 C8 57.618 20.000 2
T44 var_5 C1 C7 C8 C9 36.966 20.000 3
T44 var_6 C7 C8 N8 HN81 180.000 20.000 3
T44 var_7 C7 C8 C9 O10 -130.183 20.000 3
T44 var_8 C8 C9 O10 HO1 180.000 20.000 1
loop_
_chem_comp_chir.comp_id
_chem_comp_chir.id
_chem_comp_chir.atom_id_centre
_chem_comp_chir.atom_id_1
_chem_comp_chir.atom_id_2
_chem_comp_chir.atom_id_3
_chem_comp_chir.volume_sign
T44 chir_01 C8 C7 C9 N8 negativ
loop_
_chem_comp_plane_atom.comp_id
_chem_comp_plane_atom.plane_id
_chem_comp_plane_atom.atom_id
_chem_comp_plane_atom.dist_esd
T44 plan-1 C1 0.020
T44 plan-1 C2 0.020
T44 plan-1 C6 0.020
T44 plan-1 C7 0.020
T44 plan-1 C3 0.020
T44 plan-1 C4 0.020
T44 plan-1 C5 0.020
T44 plan-1 I3 0.020
T44 plan-1 O4 0.020
T44 plan-1 I5 0.020
T44 plan-1 H2 0.020
T44 plan-1 H6 0.020
T44 plan-2 C9 0.020
T44 plan-2 C8 0.020
T44 plan-2 O9 0.020
T44 plan-2 O10 0.020
T44 plan-3 'C1'' 0.020
T44 plan-3 'C2'' 0.020
T44 plan-3 'C6'' 0.020
T44 plan-3 O4 0.020
T44 plan-3 'C3'' 0.020
T44 plan-3 'C4'' 0.020
T44 plan-3 'C5'' 0.020
T44 plan-3 'I3'' 0.020
T44 plan-3 'O4'' 0.020
T44 plan-3 'I5'' 0.020
T44 plan-3 'H2'' 0.020
T44 plan-3 'H6'' 0.020
T44 plan-4 N8 0.020
T44 plan-4 C8 0.020
T44 plan-4 HN81 0.020
T44 plan-4 HN82 0.020
# ------------------------------------------------------
# ------------------------------------------------------
As far as I can tell this file should be correct, and I have read this
file into refmac using LIB_IN. However, during refinement the molecule
is distorted and new files describing the ligand are created in the temp
directory, which after three rounds of refinement looks like this:
global_
_lib_name mon_lib
_lib_version 3.1
_lib_update 16/07/00
# ------------------------------------------------
#
# --- LIST OF MONOMERS ---
#
data_comp_list
loop_
_chem_comp.id
_chem_comp.three_letter_code
_chem_comp.name
_chem_comp.group
_chem_comp.number_atoms_all
_chem_comp.number_atoms_nh
_chem_comp.desc_level
T44 T44 '. ' non-polymer
35 24 .
#
# --- DESCRIPTION OF MONOMERS ---
#
data_comp_T44
#
loop_
_chem_comp_atom.comp_id
_chem_comp_atom.atom_id
_chem_comp_atom.type_symbol
_chem_comp_atom.type_energy
_chem_comp_atom.partial_charge
T44 O10 O O 0.000
T44 C9 C C 0.000
T44 O9 O O 0.000
T44 C8 C CH1 0.000
T44 H8 H HCH1 0.000
T44 N8 N N 0.000
T44 C7 C CH2 0.000
T44 H71 H HCH2 0.000
T44 H72 H HCH2 0.000
T44 C1 C CR6 0.000
T44 C2 C CR16 0.000
T44 H2 H HCR6 0.000
T44 C3 C CR6 0.000
T44 I3 I I 0.000
T44 C6 C CR16 0.000
T44 H6 H HCR6 0.000
T44 C5 C CR6 0.000
T44 I5 I I 0.000
T44 C4 C CR6 0.000
T44 O4 O O2 0.000
T44 'C1'' C CH1 0.000
T44 'H6'' H HCH1 0.000
T44 'C6'' C CH1 0.000
T44 'C5'' C C 0.000
T44 'I5'' I I 0.000
T44 'C2'' C CH1 0.000
T44 'H2'' H HCH1 0.000
T44 'C3'' C C 0.000
T44 'I3'' I I 0.000
T44 'C4'' C C 0.000
T44 'O4'' O O 0.000
T44 'HO4'' H HO 0.000
T44 HN82 H HHO 0.000
T44 HN81 H HHHO 0.000
T44 HO1 H HHHH 0.000
loop_
_chem_comp_tree.comp_id
_chem_comp_tree.atom_id
_chem_comp_tree.atom_back
_chem_comp_tree.atom_forward
_chem_comp_tree.connect_type
T44 O10 n/a C9 START
T44 C9 O10 C8 .
T44 O9 C9 . .
T44 C8 C9 C7 .
T44 H8 C8 . .
T44 N8 C8 . .
T44 C7 C8 C1 .
T44 H71 C7 . .
T44 H72 C7 . .
T44 C1 C7 C6 .
T44 C2 C1 C3 .
T44 H2 C2 . .
T44 C3 C2 I3 .
T44 I3 C3 . .
T44 C6 C1 C5 .
T44 H6 C6 . .
T44 C5 C6 C4 .
T44 I5 C5 . .
T44 C4 C5 O4 .
T44 O4 C4 'C1'' .
T44 'C1'' O4 'C2'' .
T44 'H6'' 'C1'' . .
T44 'C6'' 'C1'' 'C5'' .
T44 'C5'' 'C6'' 'I5'' .
T44 'I5'' 'C5'' . .
T44 'C2'' 'C1'' 'C3'' .
T44 'H2'' 'C2'' . .
T44 'C3'' 'C2'' 'C4'' .
T44 'I3'' 'C3'' . .
T44 'C4'' 'C3'' 'O4'' .
T44 'O4'' 'C4'' 'HO4'' .
T44 'HO4'' 'O4'' HN82 .
T44 HN82 'HO4'' HN81 .
T44 HN81 HN82 HO1 .
T44 HO1 HN81 . END
T44 'C5'' 'C4'' . ADD
T44 'C6'' 'H6'' . ADD
T44 C4 C3 . ADD
loop_
_chem_comp_bond.comp_id
_chem_comp_bond.atom_id_1
_chem_comp_bond.atom_id_2
_chem_comp_bond.type
_chem_comp_bond.value_dist
_chem_comp_bond.value_dist_esd
T44 'I5'' 'C5'' . 2.065 0.020
T44 'C5'' 'C6'' . 1.500 0.020
T44 'C5'' 'C4'' . 1.330 0.020
T44 'C6'' 'H6'' . 1.099 0.020
T44 'C6'' 'C1'' . 1.524 0.020
T44 'H6'' 'C1'' . 1.099 0.020
T44 'O4'' 'C4'' . 1.220 0.020
T44 'C4'' 'C3'' . 1.330 0.020
T44 'I3'' 'C3'' . 2.065 0.020
T44 'C3'' 'C2'' . 1.500 0.020
T44 'H2'' 'C2'' . 1.099 0.020
T44 'C2'' 'C1'' . 1.524 0.020
T44 'C1'' O4 . 1.426 0.020
T44 O4 C4 . 1.370 0.020
T44 C4 C5 . 1.487 0.020
T44 C4 C3 . 1.487 0.020
T44 I5 C5 . 2.090 0.020
T44 C5 C6 . 1.390 0.020
T44 H6 C6 . 1.083 0.020
T44 C6 C1 . 1.390 0.020
T44 I3 C3 . 2.090 0.020
T44 C3 C2 . 1.390 0.020
T44 H2 C2 . 1.083 0.020
T44 C2 C1 . 1.390 0.020
T44 C1 C7 . 1.511 0.020
T44 H71 C7 . 1.092 0.020
T44 H72 C7 . 1.092 0.020
T44 C7 C8 . 1.524 0.020
T44 H8 C8 . 1.099 0.020
T44 N8 C8 . 1.455 0.020
T44 C8 C9 . 1.500 0.020
T44 O9 C9 . 1.220 0.020
T44 C9 O10 . 1.220 0.020
loop_
_chem_comp_angle.comp_id
_chem_comp_angle.atom_id_1
_chem_comp_angle.atom_id_2
_chem_comp_angle.atom_id_3
_chem_comp_angle.value_angle
_chem_comp_angle.value_angle_esd
T44 O10 C9 O9 120.000 3.000
T44 O10 C9 C8 120.500 3.000
T44 O9 C9 C8 120.500 3.000
T44 C9 C8 H8 108.810 3.000
T44 C9 C8 N8 111.600 3.000
T44 C9 C8 C7 109.470 3.000
T44 H8 C8 N8 109.470 3.000
T44 H8 C8 C7 108.340 3.000
T44 N8 C8 C7 105.000 3.000
T44 C8 C7 H71 109.470 3.000
T44 C8 C7 H72 109.470 3.000
T44 C8 C7 C1 109.470 3.000
T44 H71 C7 H72 107.900 3.000
T44 H71 C7 C1 109.470 3.000
T44 H72 C7 C1 109.470 3.000
T44 C7 C1 C2 120.000 3.000
T44 C7 C1 C6 120.000 3.000
T44 C2 C1 C6 120.000 3.000
T44 C1 C2 H2 120.000 3.000
T44 C1 C2 C3 120.000 3.000
T44 H2 C2 C3 120.000 3.000
T44 C2 C3 I3 120.000 3.000
T44 C2 C3 C4 120.000 3.000
T44 I3 C3 C4 120.000 3.000
T44 C1 C6 H6 120.000 3.000
T44 C1 C6 C5 120.000 3.000
T44 H6 C6 C5 120.000 3.000
T44 C6 C5 I5 120.000 3.000
T44 C6 C5 C4 120.000 3.000
T44 I5 C5 C4 120.000 3.000
T44 C5 C4 O4 120.000 3.000
T44 C5 C4 C3 120.000 3.000
T44 O4 C4 C3 120.000 3.000
T44 C4 O4 'C1'' 120.000 3.000
T44 O4 'C1'' 'H6'' 109.470 3.000
T44 O4 'C1'' 'C6'' 109.470 3.000
T44 O4 'C1'' 'C2'' 109.470 3.000
T44 'H6'' 'C1'' 'C6'' 108.340 3.000
T44 'H6'' 'C1'' 'C2'' 108.340 3.000
T44 'C6'' 'C1'' 'C2'' 111.000 3.000
T44 'C1'' 'H6'' 'C6'' 109.500 3.000
T44 'C1'' 'C6'' 'C5'' 109.470 3.000
T44 'C1'' 'C6'' 'H6'' 108.340 3.000
T44 'C5'' 'C6'' 'H6'' 108.810 3.000
T44 'C6'' 'C5'' 'I5'' 120.000 3.000
T44 'C6'' 'C5'' 'C4'' 120.000 3.000
T44 'I5'' 'C5'' 'C4'' 120.000 3.000
T44 'C1'' 'C2'' 'H2'' 108.340 3.000
T44 'C1'' 'C2'' 'C3'' 109.470 3.000
T44 'H2'' 'C2'' 'C3'' 108.810 3.000
T44 'C2'' 'C3'' 'I3'' 120.000 3.000
T44 'C2'' 'C3'' 'C4'' 120.000 3.000
T44 'I3'' 'C3'' 'C4'' 120.000 3.000
T44 'C3'' 'C4'' 'O4'' 120.500 3.000
T44 'C3'' 'C4'' 'C5'' 120.000 3.000
T44 'O4'' 'C4'' 'C5'' 120.500 3.000
T44 'C4'' 'O4'' 'HO4'' 120.000 3.000
loop_
_chem_comp_tor.comp_id
_chem_comp_tor.id
_chem_comp_tor.atom_id_1
_chem_comp_tor.atom_id_2
_chem_comp_tor.atom_id_3
_chem_comp_tor.atom_id_4
_chem_comp_tor.value_angle
_chem_comp_tor.value_angle_esd
_chem_comp_tor.period
T44 var_1 O10 C9 C8 C7 -131.440 20.000 3
T44 var_2 C9 C8 C7 C1 36.215 20.000 3
T44 var_3 C8 C7 C1 C6 -122.807 20.000 2
T44 CONST_1 C7 C1 C2 C3 180.000 0.000 0
T44 CONST_2 C1 C2 C3 I3 180.000 0.000 0
T44 CONST_3 C1 C2 C3 C4 0.000 0.000 0
T44 CONST_4 C7 C1 C6 C5 180.000 0.000 0
T44 CONST_5 C1 C6 C5 C4 0.000 0.000 0
T44 CONST_6 C6 C5 C4 O4 180.000 0.000 0
T44 CONST_7 C6 C5 C4 C3 0.000 0.000 0
T44 var_4 C5 C4 O4 'C1'' -101.012 20.000 1
T44 var_5 C4 O4 'C1'' 'C2'' 4.981 20.000 1
T44 var_6 O4 'C1'' 'H6'' 'C6'' 144.598 20.000 1
T44 var_7 O4 'C1'' 'C6'' 'C5'' -177.563 20.000 3
T44 var_8 O4 'C1'' 'C6'' 'H6'' -40.734 20.000 3
T44 var_9 'C1'' 'C6'' 'C5'' 'I5'' -174.209 20.000 3
T44 var_10 'C1'' 'C6'' 'C5'' 'C4'' 8.089 20.000 3
T44 var_11 O4 'C1'' 'C2'' 'C3'' 176.823 20.000 3
T44 var_12 'C1'' 'C2'' 'C3'' 'C4'' -4.834 20.000 3
T44 CONST_8 'C2'' 'C3'' 'C4'' 'O4'' 180.000 0.000 0
T44 CONST_9 'C2'' 'C3'' 'C4'' 'C5'' 0.000 0.000 0
loop_
_chem_comp_chir.comp_id
_chem_comp_chir.id
_chem_comp_chir.atom_id_centre
_chem_comp_chir.atom_id_1
_chem_comp_chir.atom_id_2
_chem_comp_chir.atom_id_3
_chem_comp_chir.volume_sign
T44 chir_01 'C1'' 'C6'' 'C2'' O4 positiv
T44 chir_02 C8 C7 N8 C9 positiv
loop_
_chem_comp_plane_atom.comp_id
_chem_comp_plane_atom.plane_id
_chem_comp_plane_atom.atom_id
_chem_comp_plane_atom.dist_esd
T44 plan-1 'C5'' 0.020
T44 plan-1 'I5'' 0.020
T44 plan-1 'C6'' 0.020
T44 plan-1 'C4'' 0.020
T44 plan-1 'O4'' 0.020
T44 plan-1 'C3'' 0.020
T44 plan-1 'I3'' 0.020
T44 plan-1 'C2'' 0.020
T44 plan-2 C4 0.020
T44 plan-2 O4 0.020
T44 plan-2 C5 0.020
T44 plan-2 C3 0.020
T44 plan-2 C6 0.020
T44 plan-2 C2 0.020
T44 plan-2 C1 0.020
T44 plan-2 I5 0.020
T44 plan-2 I3 0.020
T44 plan-2 C7 0.020
T44 plan-2 H6 0.020
T44 plan-2 H2 0.020
T44 plan-3 C9 0.020
T44 plan-3 C8 0.020
T44 plan-3 O9 0.020
T44 plan-3 O10 0.020
# ------------------------------------------------------
# ------------------------------------------------------
What is wrong, and how do I solve this problem?
Therese Eneqvist
......................
UCMP
Umeň University
SE-90187 Umeň, Sweden
tel. +46 90 7856783
fax +46 778007