Automated molecular replacement with MrBump
From Media Wiki
Main Page - Using the CCP4 software - Molecular replacement with CCP4 - Automated molecular replacement with MrBump
MrBUMP is an automated scheme for Molecular Replacement. Given a target sequence and experimental structure factors, it will search for homologous structures, create a set of suitable search models from the template structures, do molecular replacement, and test the solutions with some rounds of restrained refinement.
The scheme has the potential to be highly parallel, searching over many homologues, derived models and MR techniques. Currently it runs in two modes:
- Desktop: the scheme is streamlined for one node
- Cluster: jobs are distributed over available nodes
MrBUMP uses existing programs for the various steps. MR itself is done using Molrep or Phaser. The CCP4 suite is widely used for various steps. It also uses bioinformatics programs such as Fasta, MAFFT and ClustalW, and services provided by the European Bioinformatics Institute.
Authors: Ronan Keegan and Martyn Winn
Contents |
[edit] Running MrBump
The most common use of MrBUMP is to perform molecular replacement using just the observed structure factors and protein sequence (for other uses, see Advanced Options). To perform this calculation you must first have a set of structure factor magnitudes (usually determined as part of data processing), and the sequence.
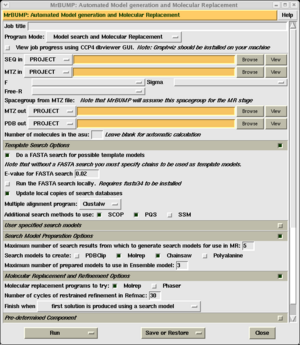
Go to the molecular replacement module from within CCP4i, and select the "MrBump" task. The MrBump task window is shown on the right.
In order to run the program, you must provide 2 files:
- A sequence file. This must contain the sequence of the target protein. The file may simply contain a list of 1-letter residue codes, or it may contain a header line (preceded with '>'), followed by the residue codes on subsequent lines (FASTA format).
- An MTZ file. This must contain the structure factor magnitudes and a column of free R flags.
If the molecular replacement calculation succeeds, the program will produce a PDB file for the MR model in the new crystal, and an MTZ file containing map coefficients. The output MTZ and PDB filenames are generated automatically, you may change them if you wish.
Select 'Run now' to start MrBump.
[edit] Program output
If a suitable solution is found, i.e. a model that refines well, the job will terminate and the final results will be displayed. The resulting refined PDB model and MTZ output from Refmac are made available to the user for further model building.
MrBUMP runs multiple jobs to try different molecular replacement methods and search models in order to try and find a solution. The individual program log files may be inspected by viewing the program output in a web browser, which will give links to the output of the individual programs.
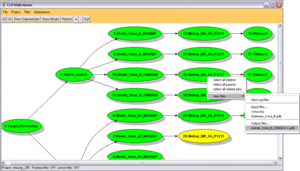
To make it easier to order to follow the progress of their MrBUMP jobs a graphical representation may used. To enable this feature, select the "View job progress using dbviewer" button at the top of the task window (note that for this feature to be made available you will need to install the Graphviz software). The viewer will show a tree of all the calculations performed so far (as shown on the right), showing all the paths explored so far in attempting to solve the structure. Any task may be selected using the mouse to view the log file.
[edit] Advanced options
- Checking your spacegroup
MrBUMP has the option to check both possibilities for an enantiomorphic spacegroup. For example, if you have spacegroup P43 then it can also check for Molecular Replacement solutions in P41.
[edit] Related pages
[edit] Program documentation
The latest version of the documentation is available here. This provides information on program keywords which may be used from the command line.
This page describes MrBump version 0.4.2 (CCP4 version 6.1.0).
--Kevin Cowtan 03:42, 22 April 2008 (CDT)