Automated experimental phasing with Crank
From Media Wiki
Main Page - Using the CCP4 software - Experimental phasing with CCP4 - Automated experimental phasing with Crank
Crank is a program to automate protein crystal structure determination, when starting from good quality reflection data. It uses many high quality crystallographic programs, giving them properly formatted input files, and interpreting their output. It is designed to be a transparent box, allowing automation of the structure determination process in easier cases, and allow the user to run and cross compare results obtained from different techniques in difficult cases.
Crank can currently use any combination of the following software as part of its automated pahsing pipeline:
- FA calculation: AFRO, SHELXC
- Heavy atom location: CRUNCH2, SHELXD
- Phasing: BP3, SHELXE
- Phase imrovement: SOLOMON, PIRATE
- Model building: BUCCANEER, ARP/WARP
In addition to running a full pipeline, a subset of tasks may be run so the user can perform the remaining steps externally.
Contents |
[edit] Running CRANK
To run CRANK, you must first have a set of experimental data, either from a SAD, MAD or SIRAS experiment. It is useful to have the sequence of the protein.
Select the 'Experimental phasing' module from with CCP4i. Select the 'Crank' task from the 'Automated search & phasing' folder. The task window should appear as shown on the right.
First, select whether you want to perform the calculation using Intesities or Magnitudes. Next select the typpe of experiment which was performed. For a single crystal anomalous scattering experiment, select SAD, 2WMAD, 3WMAD or 4WMAD, depending on the number of wavelengths used. If a native and isomorphous derivative were used, select SIRAS, 2WMADN, 3WMADN or 4WMADN.
Next you should normally provide 2 files:
- An MTZ file. This must contain the stucture factor intensities or magnitudes for all wavelengths and/or derivatives, in a single file. If your data are in separate files, these should first be merged.
- A sequence file. This must contain the sequence of the protein. The file may simply contain a list of 1-letter residue codes, or it may contain multiple chains, each specified by a chain ID (preceded with '>'), followed by the residue codes on subsequent lines (FASTA format).
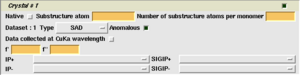
The task window will now display a section for each dataset - one section for each wavelength, and if required an initial section for the native dataset. For the native dataset, you must provide the MTZ column names for the oberserved magitude or intensity and its standard deviation.
For each wavelength, you must then supply the expected anomalous scatterer type, a rough guess at the number of anomalous scatterers in the assymentric unit, and the type of dataset - peak, inflection, remote etc. You must also supply the expected anomalous scattering coefficients - f' and f" - for this atom type at the given wavelength (these are obtained by fluorescence scan or from the wavelength: [1], [2]). Alternatively, if the data was collected with Cu Kα radiation, simply check the box. Finally select the column labels for the observed magitudes or intensities and their standard deviations.
Finally, you can choose which software to use to solve your structure. The options include:
- CRUNCH2/BP3/SOLOMON/ARPWARP - robust pipeline for difficult problems.
- SHELXCDE/BP3/SOLOMON/ARPWARP - faster pipeline using SHELX for heavy atom location.
- SHELXCDE/ARPWARP - faster still, but dependent on good data.
- CRUNCH2/BP3/SOLOMON/PIRATE/BUCCANEER - pipeline using only CCP4 tools - no external dependencies.
- CUSTOM - design your own pipeline.
Select 'Run now' to start Crank.
[edit] Program output
[edit] Output MTZ file
In the Crank CCP4i interface, you specify the Crank output MTZ file, this file is created in the directory you specify. If you have troubles finding it, look in your CCP4 project directories. This .mtz file contains the final structure factors and Hendrickson-Lattman parameters, as estimated by the various programs in the crystallographic pipeline.
[edit] Crank log file
The Crank log file contains output not only from Crank, but from all the programs in the pipeline, therefore, this file can grow very large. Although we apologize for the size, we think that it is important for the user to read and understand the information in the logfile.
To this end, we are currently developing tools in the Crank suite to parse the logfile information from all the various crystallographic tools and to convert it into a format amenable both for users to examine and for programs to use to make decisions on the direction of the pipeline, depending on program data.
Until then, some hints about what to look for in the logfile:
- Crank header and references. First, Crank outputs its header and a customized reference section to use when writing a paper. It then outputs the XML that was input into it, to allow for easier debugging of program operation.
- Individual Program Logfiles. In addition to these standard Crank logfiles, depending on the programs that were invoked by Crank, additional sections will be appended to the logfile for each program that is run. In the next section we will talk about the program output of the crystallographic programs in Crank.
[edit] Advanced options
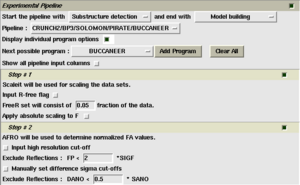
If you wish to run only part of the pipeline, then select the start and endpoint for the calculation using the first two menus in the 'Experimental pipeline' section of the task window, shown on the right.
For fine grained control of individual tasks in the pipeline, select the 'Display individual program options checkbox. A set of program options will appear for each step in the pipeline, as shown on the right.
[edit] Related pages
[edit] Program documentation
The latest version of the documentation is available here. This provides information on program keywords which may be used from the command line.
This page describes CRANK version 1.1 (CCP4 version 6.1.0).