Contents
IntroductionSecondary Structure (SSM)
Close Atoms
Selected residues
Transforming Coordinates
Saving the transformed coordinates
Methodology
 |
CCP4 Molecular Graphics Documentation | |
| Superpose |
| Documentation Contents | On-line Documentation | Tutorials | CCP4mg Home |
Access to the Superpose application is from the Application menu. There are four different methods for superposing structures. They are explained in the folllowing sections. The method can be chosen from the menu at the top of the Superpose window.
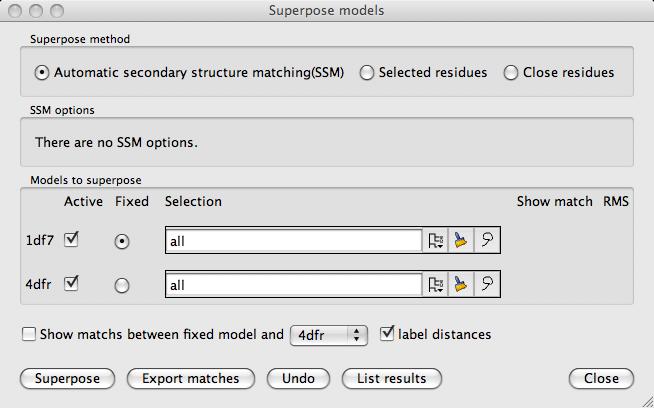
All the loaded models are listed, one per line, in the window. On the line for each model is:
A check button to indicate if the model is to be superposed (by default is on)
A radio button to indicate if this model is the fixed model
A selection widget to select a limited set of atoms to be used in the superposition (default 'All')
Click the button labelled Superpose to superpose the structures.
It is usually easier to see the resultant superposition if the options on the main
Molecule pull-down
menu are used to set Selection.. to CA trace and
Colour.. to By molecule.
All of the molecules are moved to the position of one fixed 'target' molecule which does not move.
After superposition, if Show matched atoms is set on, the equivalent atoms in the target model and
one other selected model can be indicated by dashed lines between the atoms.
The inter-atomic distances are also shown.
After the structures are superposed the window is updated to include buttons to Undo
the superposition and Save coordinates to file.
The List results will give a listing of the matched SSEs (secondary structure elements) and residues, the inter-atomic distances and the transformation matrix. In the listing window there is an option to Save to file.
It is possible to limit the region of the structure that is superposed using the selection menu which is, by default, labelled All atoms. For some cases it may only be necessary to select the limited set of atoms for the target model - this is discussed further for each method below.
This method will superpose protein structures very quickly and without needing any input from the user to specify equivalent residues. The method works equally well for close homologs or for matching limited regions of similarity in very different structures. The user can select limited regions of the loaded models for matching but note that the method works by first matching equivalent secondary structure elements so is not applicable to very small fragments of protein or non-protein which do not include a α-helix or β-strand. If the user's selected atoms include one Cα from a SSE then the whole of the SSE will be used in the superposition.
For some structures there may be several possible superpositions which score similarly so there is a an option to show other possible matches for all superposed molecules.
This method is useful for performing locally optimised superposition after a global superposition by SSM. For example this method will optimise the superposition of the residues forming the binding site of a ligand. To do this you need to select the required local region of the 'fixed' model. This is probably best done using the Neighbourhood of.. option from the 'fixed' model's selection menu and selecting the Neighbourhood of the ligand. It is not necessary to make any atom selection for the 'moving' models.
This method will, by default, do an SSM superposition to get the models reasonably superposed. It will then attempt to find the equivalent residues in the 'moving' model to the selected residues in the 'fixed' model. The criteria for equivalent residues is that the Cα atoms are within a given cutoff distance and the residues have similar orientation. The equivalent atoms within the equivalent residues must have the same name and be within a given cutoff distance. A least squares fit of all equivalent atoms is performed.
This method is closest in principle to the CCP4 program LSQKAB and will superpose proteins or nucleic acids.
You must select the residues to superpose for each of the structures using the selection widget. You will be warned if you have not selected an equal number of residues for each structure. By default the CA atoms of selected residues are superposed but the menu labelled For range selection match .. atoms allows alternative selections.
You can enter a transformation matrix or move the model manually: from the model icon menu select Transform coordinates and then choose one of the options..
In the window you can see the rotation matrix and translation vector that are currently applied to the coordinates and can reset these before clcking the Apply button to apply the transformation. Beware that you may need to click the Undo button to undo any existing transformation. The program checks that your input has the correct number of elements and that the rotation matrix has a determinant of one.
The model is made the 'active' moving object which is indicated by the model icon being highlighted in colour gold. To move the model you should hold down the Ctrl key and use either the left mouse button to rotate or the middle mouse button to translate the model. By default the rotation is about the centre of the model but you can reset the centre of rotation to an atom, residue, secondary structure element or chain by clicking on an atom in the model with the right mouse button and selecting Rotate around this from the pop-up menu.
This will return the model to its initial position.
The model will remain in the transformed position until the transformation is undone but the new coordinates will not be saved to file unless you explicitly do so. The transformation will be saved between program sessions and in status files and will be reapplied when the model is loaded. The new coordinates can be saved to file by selecting the File save/restore option from the model icon menu.
The Secondary Structure (SSM) superposition method superposes pairs of structures by the steps:
Find the secondary structure elements (SSEs) and represent them as one simple vector spanning
the length of the SSE
Find equivalent SSEs in the two structures using graph-theory matching by geometric criteria of distances and angles between the vectors
Superpose vectors representing equivalent SSEs
Find the most likely equivalent residues in the superposed SSEs
Superpose CA atoms of equivalent residues
Iterate the last two steps
The same procedure is also available on the EBI web server http://www.ebi.ac.uk/msd-srv/ssm
Krissinel & K. Hendrick (2004). Proceedings of CCP4 Study Weekend. Acta Cryst. D.
E.M. Mitchell et.al. (1989). J. Mol. Biol. 212 151-166